|
|
Examples of Search Inputs
How to use HD-RNAS
Search by Organism
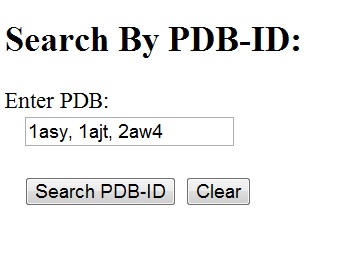
Search by PDB-ID
PDB search can be done either from the HD-RNAS homepage or the 'Search' page. In both cases, one or more PDB-IDs should be entered in the given search box, separated by commas(,). Blank spaces are optional. All the chains present in the PDB(s) are displayed in the result page. Links to their corresponding classes are also given. Chain identifier or suffixes after the PDB-ID are not accepted. Search by Sequence
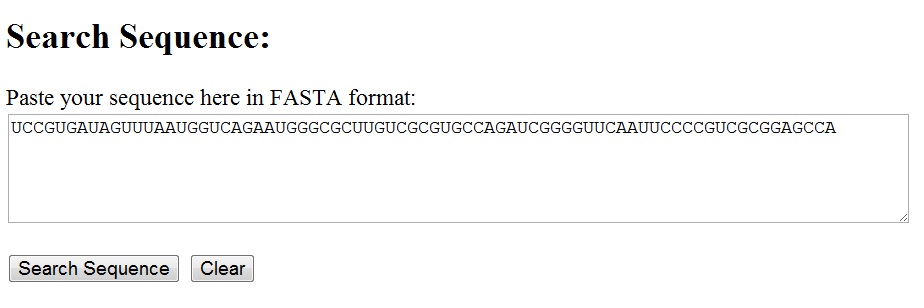
Any nucleotide sequence can be entered into the text area for the sequence search utility. The sequence can be in free format or fasta format, with or without the identifier line starting with '>' [see below]. The search is cas-insensitive.
For 'Organism Search', the scientific name of the desired organism should be entered, either in full form or abbreviated form (see below). The input is case-insensitive. The database does not support use of common names.
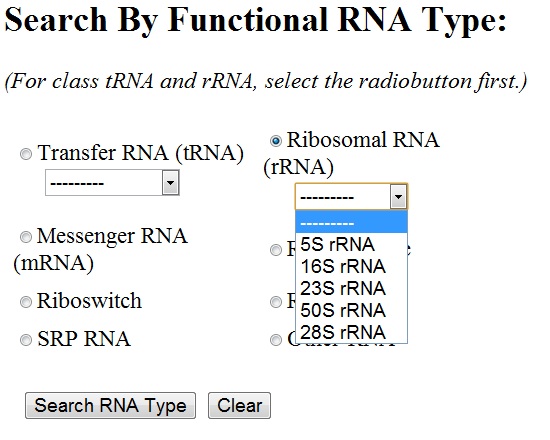
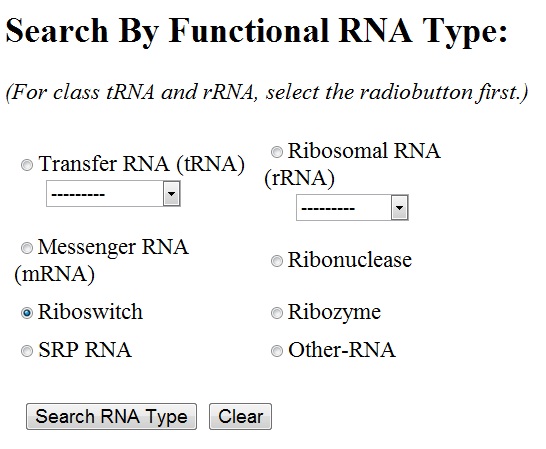
You can select any of the functional RNA type by clicking the radiobutton next to it. For ribosomal RNA and transfer RNA, sub-types are to be chosen from the drop-down menu. By default, all the sub-types of rRNA/tRNA are chosen.

'Advanced Search' page allows you to join your search criteria together. The search inputs are similar to their individual search utilities. However, the users can add the resolution of a structure as well as release date to their search options [see below].